Aethylendiamintetraacetat | 32446-51-8
-
物化性质
-
计算性质
-
ADMET
-
安全信息
-
SDS
-
制备方法与用途
-
上下游信息
-
文献信息
-
表征谱图
-
同类化合物
-
相关功能分类
-
相关结构分类
计算性质
-
辛醇/水分配系数(LogP):-4.6
-
重原子数:20
-
可旋转键数:9
-
环数:0.0
-
sp3杂化的碳原子比例:0.6
-
拓扑面积:161
-
氢给体数:2
-
氢受体数:10
反应信息
-
作为反应物:描述:Aethylendiamintetraacetat 在 氢化锶 作用下, 以 水 为溶剂, 生成 strontium ethylenediaminetetraacetate 、 alkaline earth salt of/the/ methylsulfuric acid参考文献:名称:EDTA 水溶液系统的热力学:表观和偏摩尔热容以及 EDTA 锶和钡水溶液的体积摘要:已确定 Na2SrEDTA (aq) 和 Na2BaEDTA (aq) 的表观摩尔热容和体积。已计算标准状态偏摩尔热容和体积,以及各种热力学计算所需的 0.1 m 离子强度下的偏摩尔特性。从文献中选择的焓和稳定性常数已与热容相结合,以生成 200°C 的预测稳定性常数。DOI:10.1007/bf00645193
-
作为产物:描述:strontium ethylenediaminetetraacetate 在 氢气 作用下, 以 水 为溶剂, 生成 Aethylendiamintetraacetat 、 alkaline earth salt of/the/ methylsulfuric acid参考文献:名称:EDTA 水溶液系统的热力学:表观和偏摩尔热容以及 EDTA 锶和钡水溶液的体积摘要:已确定 Na2SrEDTA (aq) 和 Na2BaEDTA (aq) 的表观摩尔热容和体积。已计算标准状态偏摩尔热容和体积,以及各种热力学计算所需的 0.1 m 离子强度下的偏摩尔特性。从文献中选择的焓和稳定性常数已与热容相结合,以生成 200°C 的预测稳定性常数。DOI:10.1007/bf00645193
文献信息
-
Cloning, Sequencing, and Characterization of a Gene Cluster Involved in EDTA Degradation from the Bacterium BNC1作者:Jan Bohuslavek、Jason W. Payne、Yong Liu、Harvey Bolton、Luying XunDOI:10.1128/aem.67.2.688-695.2001日期:2001.2
ABSTRACT EDTA is a chelating agent, widely used in many industries. Because of its ability to mobilize heavy metals and radionuclides, it can be an environmental pollutant. The EDTA monooxygenases that initiate EDTA degradation have been purified and characterized in bacterial strains BNC1 and DSM 9103. However, the genes encoding the enzymes have not been reported. The EDTA monooxygenase gene was cloned by probing a genomic library of strain BNC1 with a probe generated from the N-terminal amino acid sequence of the monooxygenase. Sequencing of the cloned DNA fragment revealed a gene cluster containing eight genes. Two of the genes,
emoA andemoB , were expressed inEscherichia coli , and the gene products, EmoA and EmoB, were purified and characterized. Both experimental data and sequence analysis showed that EmoA is a reduced flavin mononucleotide-utilizing monooxygenase and that EmoB is an NADH:flavin mononucleotide oxidoreductase. The two-enzyme system oxidized EDTA to ethylenediaminediacetate (EDDA) and nitrilotriacetate (NTA) to iminodiacetate (IDA) with the production of glyoxylate. TheemoA andemoB genes were cotranscribed when BNC1 cells were grown on EDTA. Other genes in the cluster encoded a hypothetical transport system, a putative regulatory protein, and IDA oxidase that oxidizes IDA and EDDA. We concluded that this gene cluster is responsible for the initial steps of EDTA and NTA degradation.摘要 EDTA 是一种螯合剂,广泛应用于许多行业。由于乙二胺四乙酸具有移动重金属和放射性核素的能力,因此可能成为一种环境污染物。已经在细菌菌株 BNC1 和 DSM 9103 中纯化并鉴定了启动 EDTA 降解的 EDTA 单氧化酶。然而,编码这些酶的基因尚未见报道。通过对菌株 BNC1 的基因组文库进行探针探测,克隆了 EDTA 单加氧酶基因,该探针由单加氧酶 N 端氨基酸序列生成。对克隆的 DNA 片段进行测序后发现,该基因簇包含 8 个基因。其中两个基因 emoA 和 emoB 在 大肠杆菌中表达 中表达,并纯化和鉴定了基因产物 EmoA 和 EmoB。实验数据和序列分析表明,EmoA 是一种还原型黄素单核苷酸利用单氧化酶,EmoB 是一种 NADH:黄素单核苷酸氧化还原酶。双酶系统将乙二胺四乙酸(EDTA)氧化成乙二胺二乙酸酯(EDDA),将亚硝基三乙酸酯(NTA)氧化成亚胺二乙酸酯(IDA),并产生乙醛酸。该 emoA 和 基因 基因被共转录。该基因簇中的其他基因编码一个假定的运输系统、一个假定的调控蛋白以及氧化 IDA 和 EDDA 的 IDA 氧化酶。我们的结论是,该基因簇负责 EDTA 和 NTA 降解的初始步骤。 -
Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103作者:M Witschel、S Nagel、T EgliDOI:10.1128/jb.179.22.6937-6943.1997日期:1997.11
In a gram-negative isolate (DSM 9103) able to grow with EDTA as the sole source of carbon, nitrogen, and energy, the first two steps of the catabolic pathway for EDTA were elucidated. They consisted of the sequential oxidative removal of two acetyl groups, resulting in the formation of glyoxylate. An enzyme complex that catalyzes the removal of two acetyl groups was purified and characterized. In the reaction, ethylenediaminetriacetate (ED3A) was formed as an intermediate and N,N'-ethylenediaminediacetate was the end product. The enzyme complex consisted of two components: component A' (cA'), most likely a monooxygenase, which catalyzes the cleavage of EDTA and ED3A while consuming oxygen and reduced flavin mononucleotide (FMN)-H2, and component B' (cB'), an NADH2:FMN oxidoreductase that provides FMNH2 for cA'. cB' could be replaced by other NADH2:FMN oxidoreductases such as component B of the nitrilotriacetate monooxygenase or the NADH2:FMN oxidoreductase from Photobacterium fischeri. The EDTA-oxidizing enzyme complex accepted EDTA as a substrate only when it was complexed with Mg2+, Zn2+, Mn2+, Co2+, or Cu2+. Moreover, the enzyme complex catalyzed the removal of acetyl groups from several other aminopolycarboxylic acids that possess three or more acetyl groups.
在一种革兰氏阴性分离物(DSM 9103)中,能够使用EDTA作为唯一的碳、氮和能源来源进行生长,揭示了EDTA的分解途径的前两个步骤。它们包括连续氧化去除两个乙酰基,形成乙醛酸。纯化和表征了一个催化去除两个乙酰基的酶复合物。在反应中,乙二胺四乙酸(ED3A)被形成为一种中间体,而N,N'-乙二胺二乙酸是最终产物。酶复合物由两个组分组成:组分A'(cA'),很可能是单加氧酶,催化EDTA和ED3A的裂解,同时消耗氧和还原的黄素单核苷酸(FMN)-H2,组分B'(cB'),一种NADH2:FMN氧化还原酶,为cA'提供FMNH2。cB'可以被其他NADH2:FMN氧化还原酶所取代,例如nitrilotriacetate单加氧酶的组分B或来自Photobacterium fischeri的NADH2:FMN氧化还原酶。EDTA氧化酶复合物仅在其与Mg2+,Zn2+,Mn2+,Co2+或Cu2+形成配合物时才接受EDTA作为底物。此外,酶复合物催化从几种具有三个或更多乙酰基的氨基聚羧酸中去除乙酰基。 -
Purification and Characterization of EDTA Monooxygenase from the EDTA-Degrading Bacterium BNC1作者:Jason W. Payne、Harvey Bolton、James A. Campbell、Luying XunDOI:10.1128/jb.180.15.3823-3827.1998日期:1998.8
ABSTRACT The synthetic chelating agent EDTA can mobilize radionuclides and heavy metals in the environment. Biodegradation of EDTA should reduce this mobilization. Although several bacteria have been reported to mineralize EDTA, little is known about the biochemistry of EDTA degradation. Understanding the biochemistry will facilitate the removal of EDTA from the environment. EDTA-degrading activities were detected in cell extracts of bacterium BNC1 when flavin mononucleotide (FMN), NADH, and O 2 were present. The degradative enzyme system was separated into two different enzymes, EDTA monooxygenase and an FMN reductase. EDTA monooxygenase oxidized EDTA to glyoxylate and ethylenediaminetriacetate (ED3A), with the coconsumption of FMNH 2 and O 2 . The FMN reductase provided EDTA monooxygenase with FMNH 2 by reducing FMN with NADH. The FMN reductase was successfully substituted in the assay mixture by other FMN reductases. EDTA monooxygenase was purified to greater than 95% homogeneity and had a single polypeptide with a molecular weight of 45,000. The enzyme oxidized both EDTA complexed with various metal ions and uncomplexed EDTA. The optimal conditions for activity were pH 7.8 and 35°C.
K m s were 34.1 μM for uncomplexed EDTA and 8.5 μM for MgEDTA 2− ; this difference inK m indicates that the enzyme has greater affinity for MgEDTA 2− . The enzyme also catalyzed the release of glyoxylate from nitrilotriacetate and diethylenetriaminepentaacetate. EDTA monooxygenase belongs to a small group of FMNH 2 -utilizing monooxygenases that attack carbon-nitrogen, carbon-sulfur, and carbon-carbon double bonds.摘要 合成螯合剂 EDTA 可迁移环境中的放射性核素和重金属。EDTA 的生物降解应能减少这种迁移。虽然有报道称几种细菌能使 EDTA 矿化,但人们对 EDTA 降解的生物化学过程知之甚少。了解其生物化学过程将有助于从环境中清除 EDTA。当黄素单核苷酸(FMN)、NADH 和 O 2 存在时,在 BNC1 细菌的细胞提取物中检测到 EDTA 降解活性。降解酶系统分为两种不同的酶,即 EDTA 单氧化酶和 FMN 还原酶。乙二胺四乙酸单加氧酶将乙二胺四乙酸氧化成乙醛酸和乙二胺三乙酸(ED3A),同时消耗 FMNH 2 和 O 2 .FMN 还原酶为 EDTA 单加氧酶提供 FMNH 2 通过用 NADH 还原 FMN,为 EDTA 单氧化酶提供 FMNH 2。FMN 还原酶成功地被其他 FMN 还原酶取代。乙二胺四乙酸单加氧酶的纯度超过 95%,分子量为 45,000 的单多肽。该酶既能氧化与各种金属离子络合的 EDTA,也能氧化未络合的 EDTA。活性的最佳条件是 pH 值为 7.8,温度为 35°C。 K m 为 34.1 μM,MgEDTA 2- 的 K m s 为 34.1 μM,而 MgEDTA 2- 为 8.5 μM。 K m 表明该酶对 MgEDTA 2- .该酶还能催化三醋酸氮和五醋酸二乙烯三胺释放乙醛酸。乙二胺四乙酸单加氧酶属于一小群 FMNH 2 -利用的单加氧酶,可攻击碳氮、碳硫和碳碳双键。 -
Evans, Dennis F.; Jakubovic, David A., Journal of the Chemical Society, Dalton Transactions, 1988, p. 2927 - 2934作者:Evans, Dennis F.、Jakubovic, David A.DOI:——日期:——
表征谱图
-
氢谱1HNMR
-
质谱MS
-
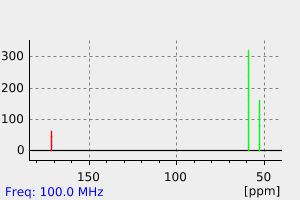
碳谱13CNMR
-
红外IR
-
拉曼Raman
-
峰位数据
-
峰位匹配
-
表征信息